Valuable data buried in journals
More than 15 years after Moneyball revolutionized how baseball teams manage their players, the practice of applying data analytics to optimize complex processes is rapidly changing many industries, including materials. However, informative machine learning (ML)–based models require clean, well-structured training datasets. Unfortunately for materials scientists, vast troves of valuable property data remain in non-machine-readable formats such as journal articles, figures, and published tables.
Collaborating to extract MPEA data
Combining specialized software tools developed by Citrine and domain knowledge from collaborators at the University of California Santa Barbara (UCSB), the French National Centre for Scientific Research (CNRS), and the United States Air Force Research Laboratory (AFRL), we developed a pipeline (shown below) to ingest, analyze, and verify a database of mechanical properties of multi-principal element alloys (MPEAs), sometimes referred to as high-entropy alloys (HEAs).
Publication
The database resulting from this work is publicly available and periodically updated on the Citrination Platform hosted by Citrine. The work has been published in Nature. Nature Scientific Data publication.
DATA INGESTION PIPELINE
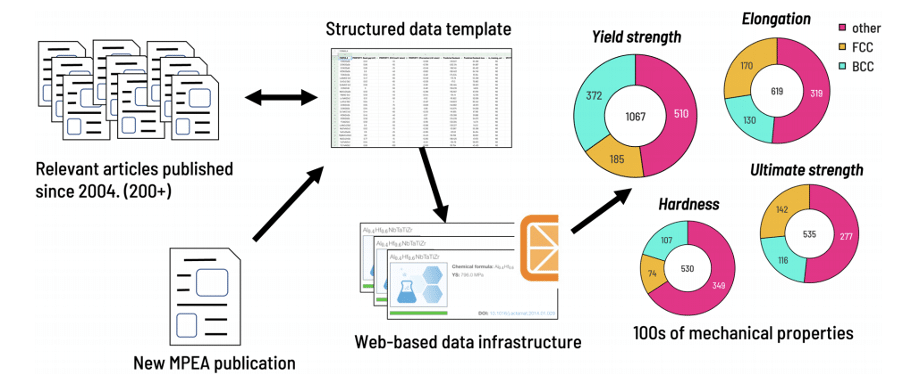
- Find relevant recent research articles and previously published reviews
- Structure data in a template according to a predefined schema
- Process aggregated data and checked for errors utilizing a combination of specialized software tools and web-based data infrastructure
PROPERTIES OF INTEREST
We focused our data capture on MPEA composition, phase stability, and mechanical properties.
QUALITY CHECKING
To help ensure data quality, a suite of Python scripts subjected each data point to a multi-step verification process.
A few of the steps are highlighted below:
- The data type of properties expecting real numbers were validated. For example, approximations were sanitized (“~250” → “250”) . Typos were highlighted and corrected or thrown away.
- Valid chemical compositions were verified via the Pymatgen library and then normalized and alphabetized.
- Processing method was assigned to one of five categories: CAST, ANNEAL, WROUGHT, POWDER, OTHER.
- Duplicate records were identified by various groupings of DOI, composition, and processing.
- Outliers were identified via ML-based cross-validation.
These steps ensured that each data point was easily traceable to the original report and that any transcription errors would be minimized.
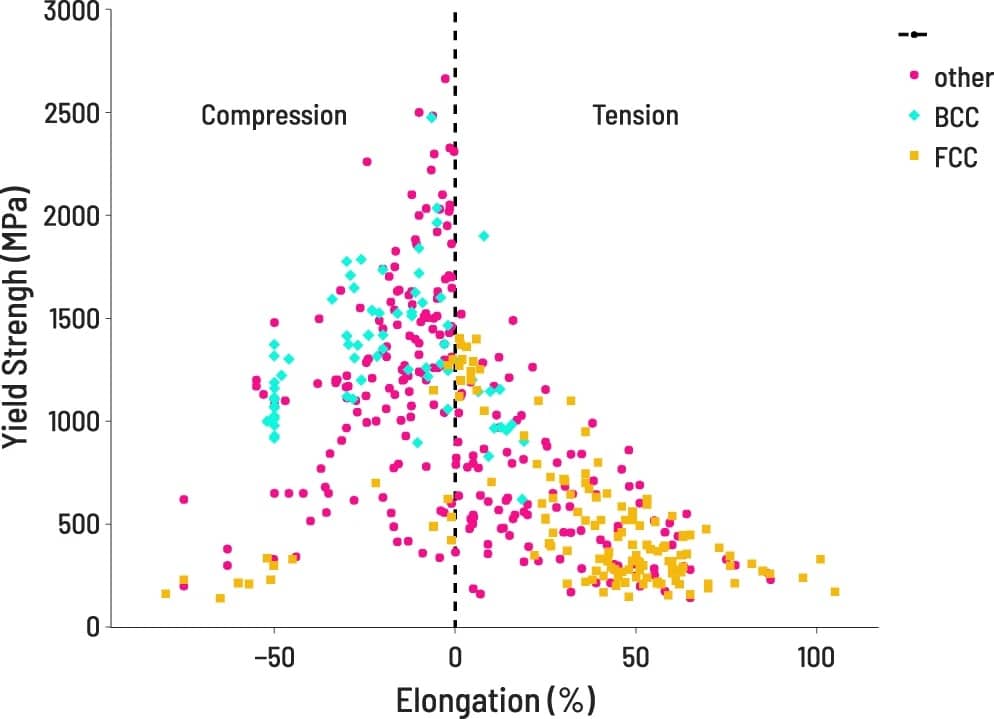
VISUALIZATION
After processing, properties of interest are easily accessible for visualization.
Feedback
We welcome any comments, feedback, or contributions to this MPEA dataset. Get in touch with Chris Borg.